v19.04
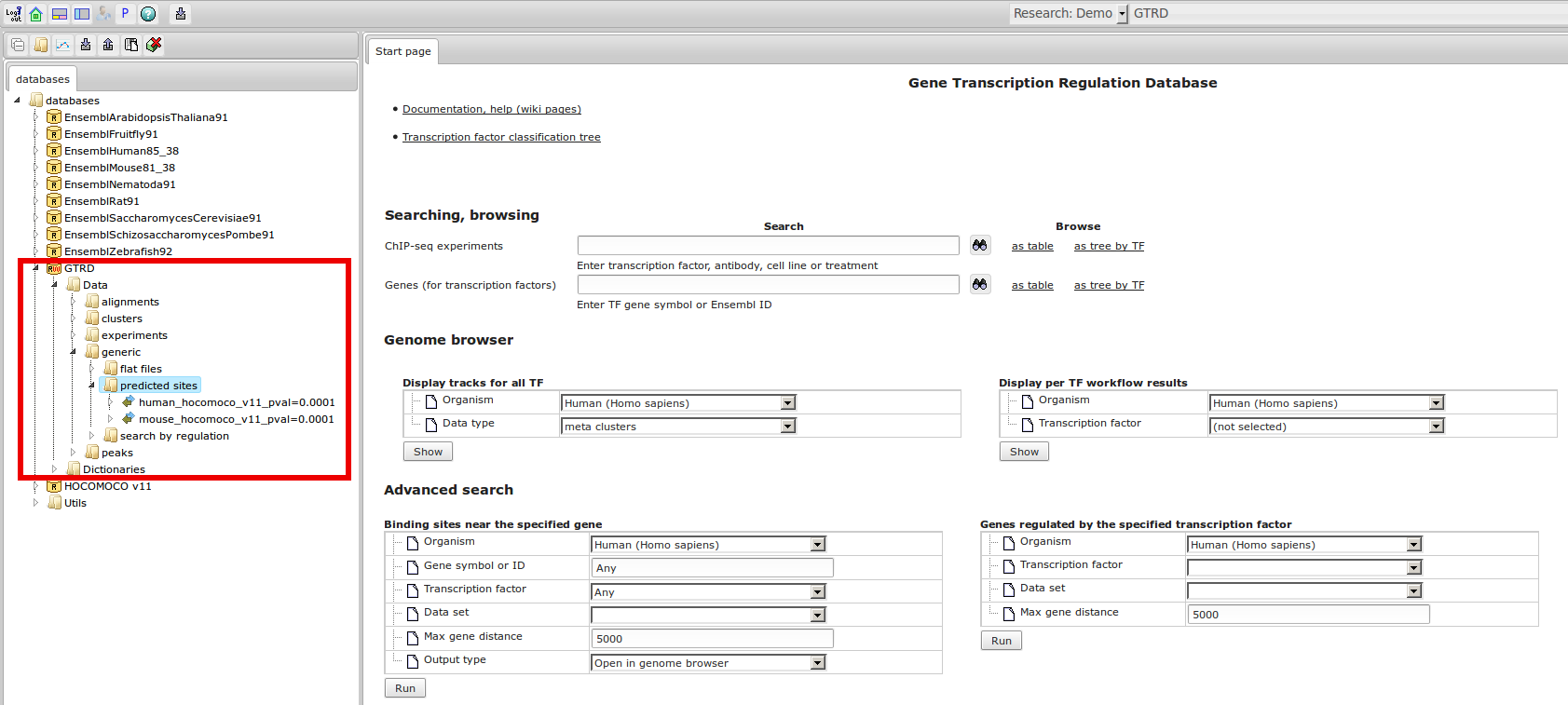
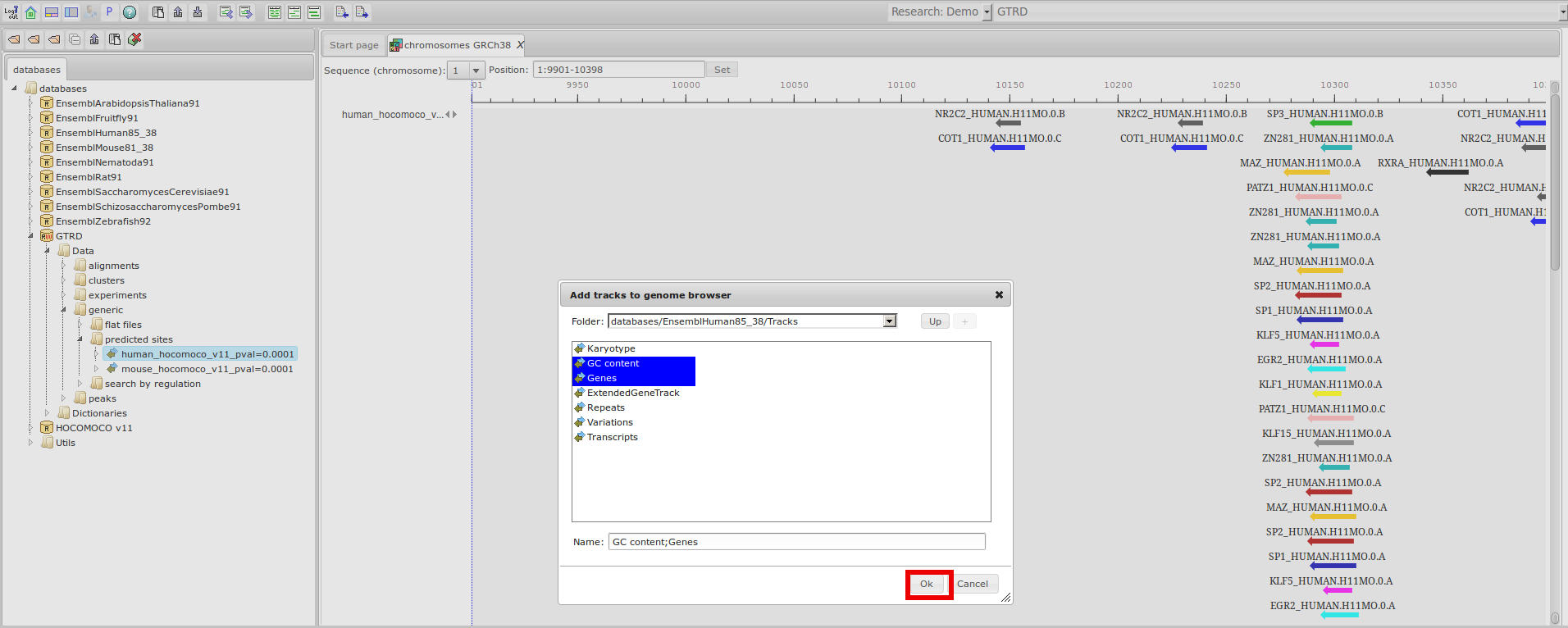
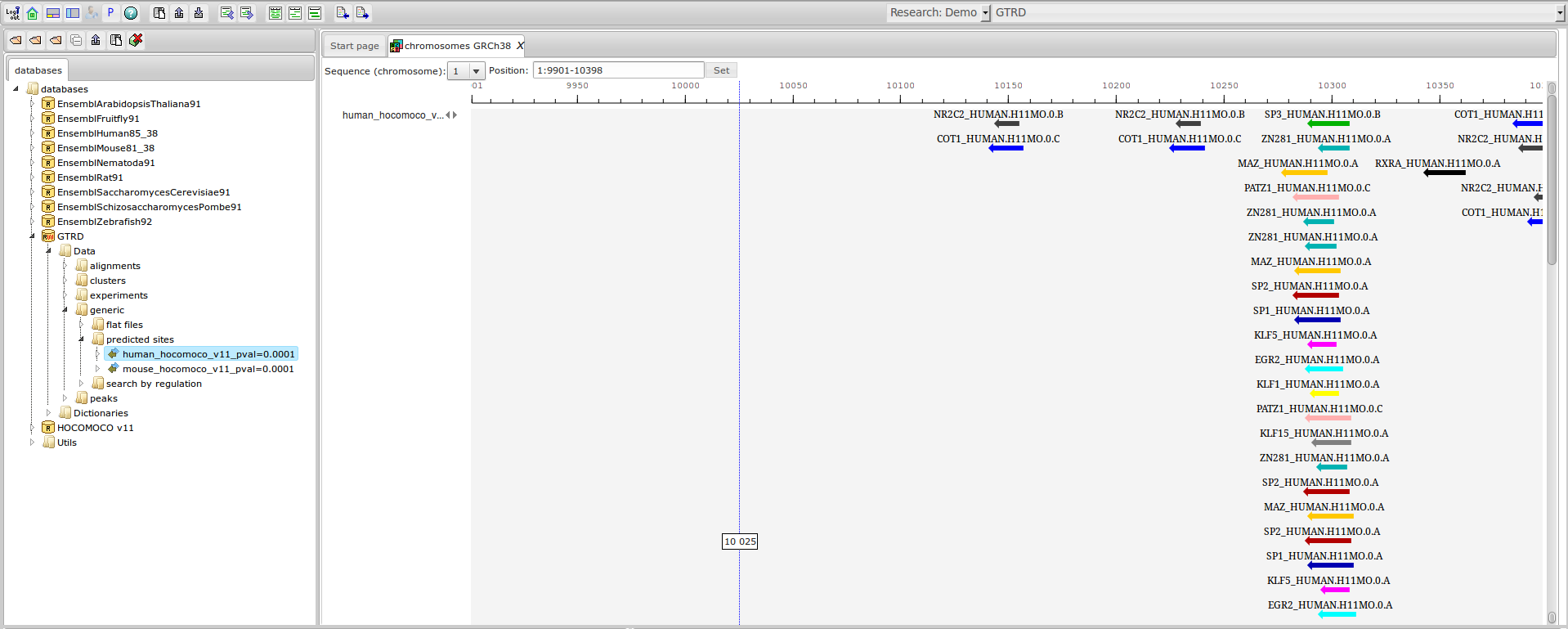
GTRD
Gene Transcription Regulation Database
The most complete collection of uniformly processed ChIP-seq data to identify transcription factor binding sites for human and mouse. Convenient web interface with advanced search, browsing and genome browser based on the BioUML platform. For support or any questions contact ivan@dote.ru
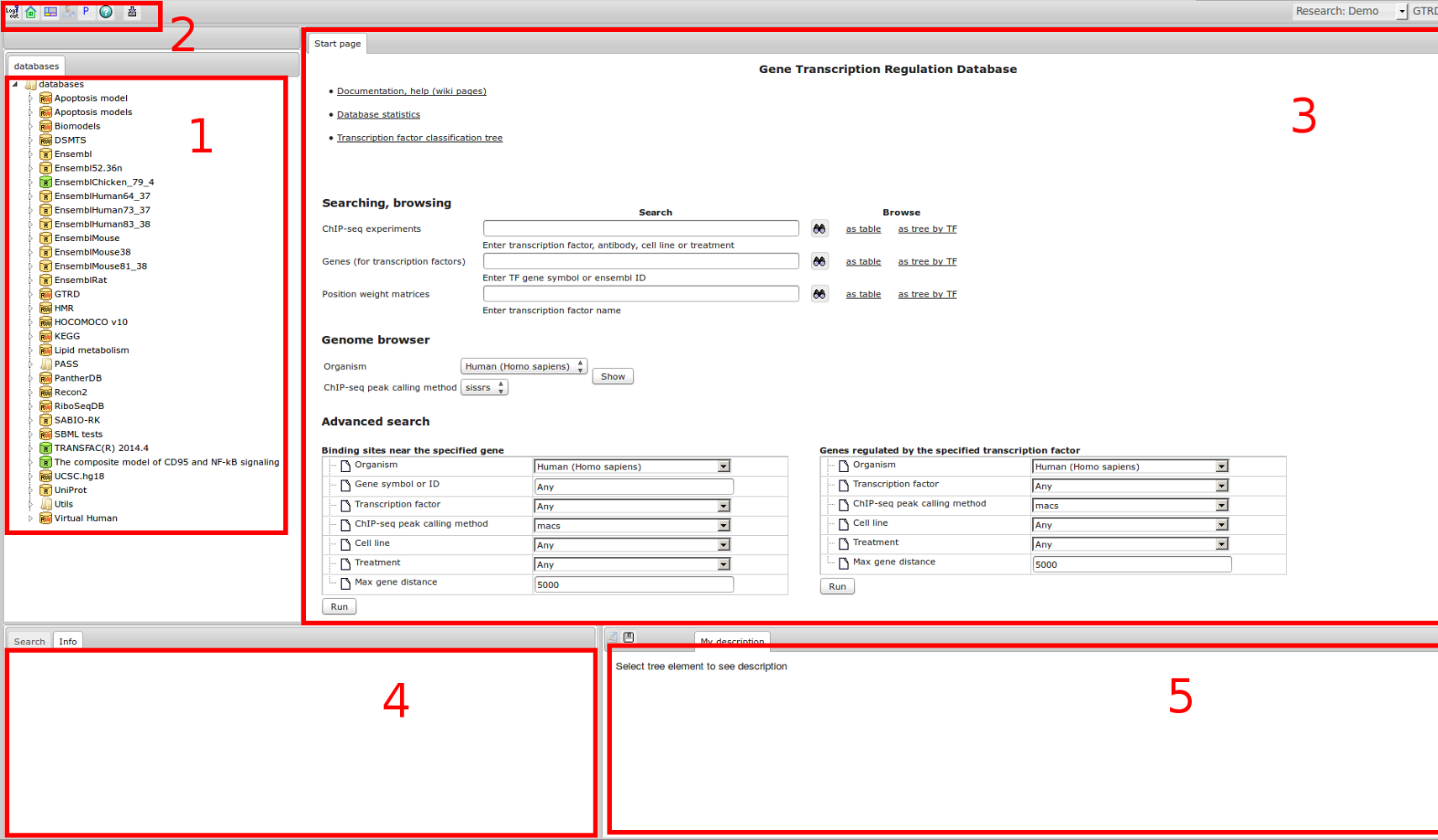
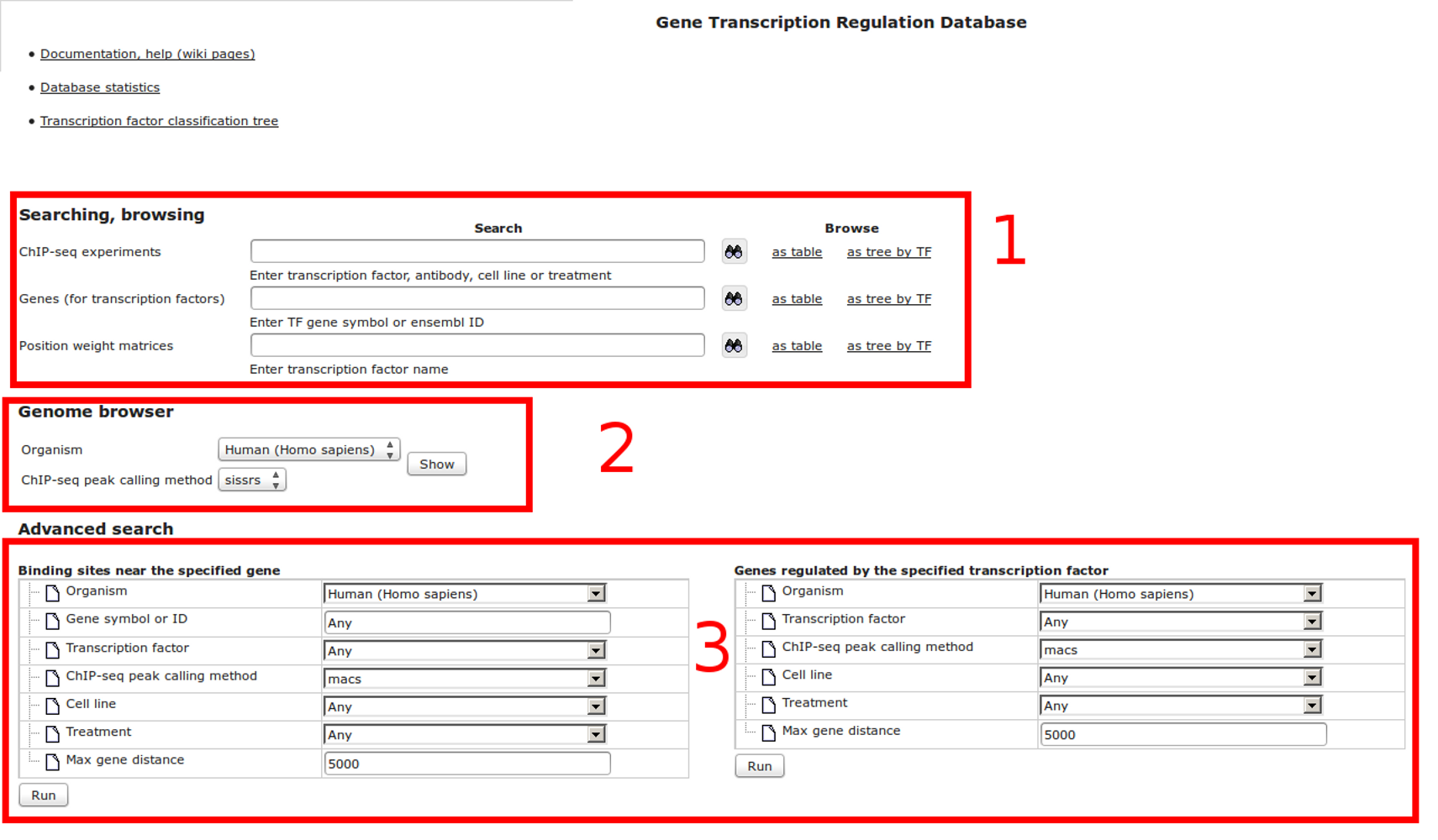
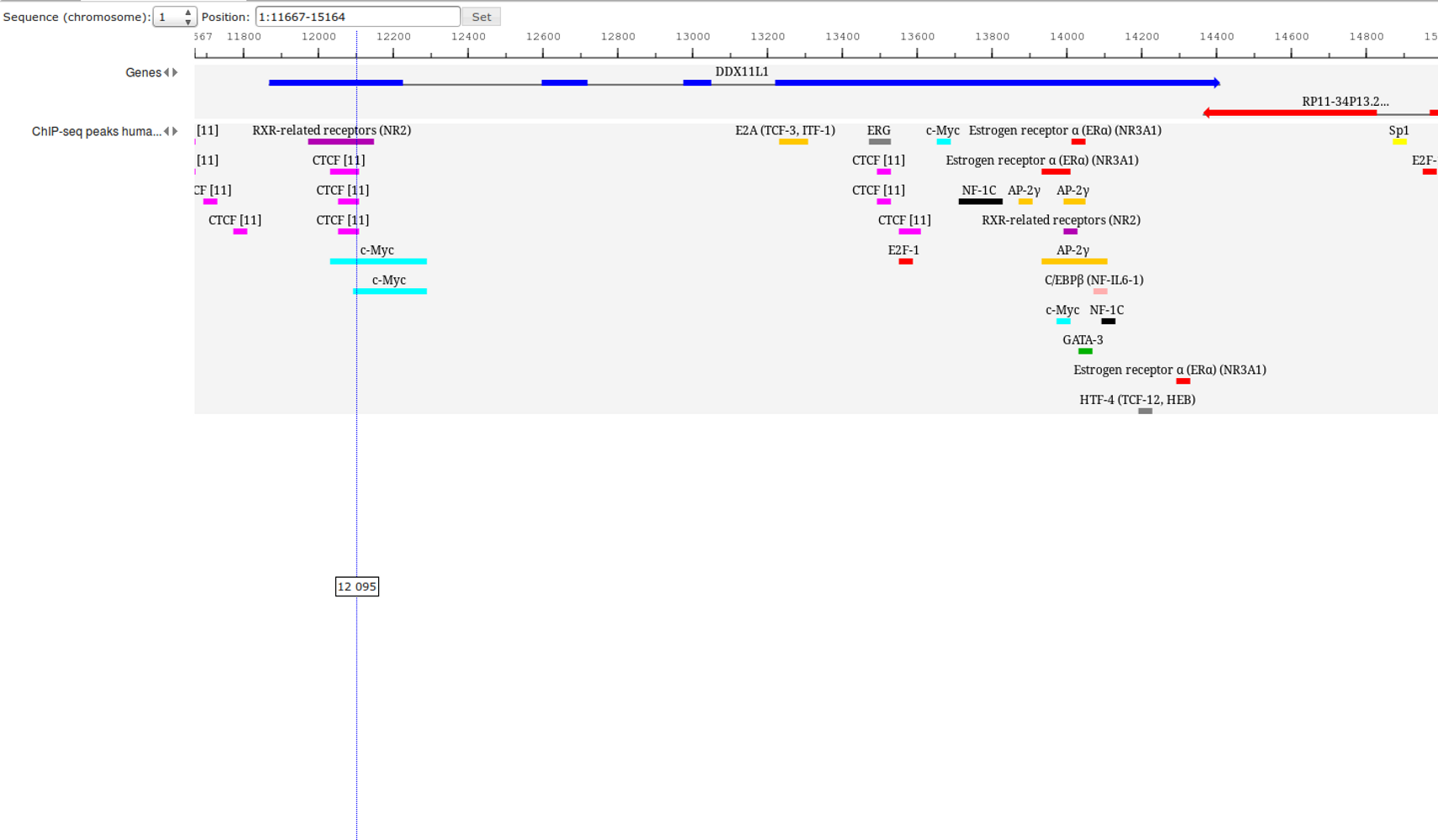
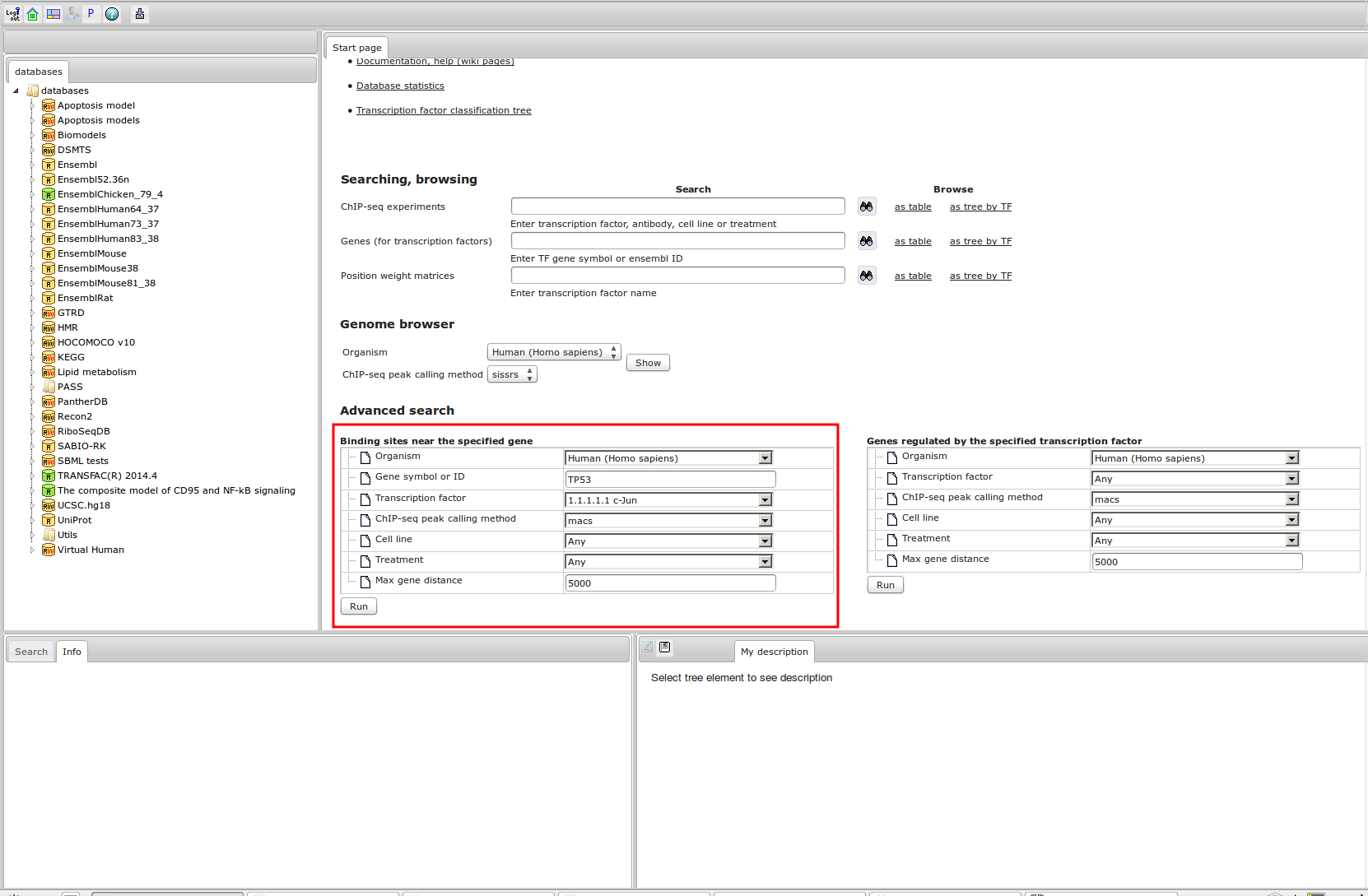
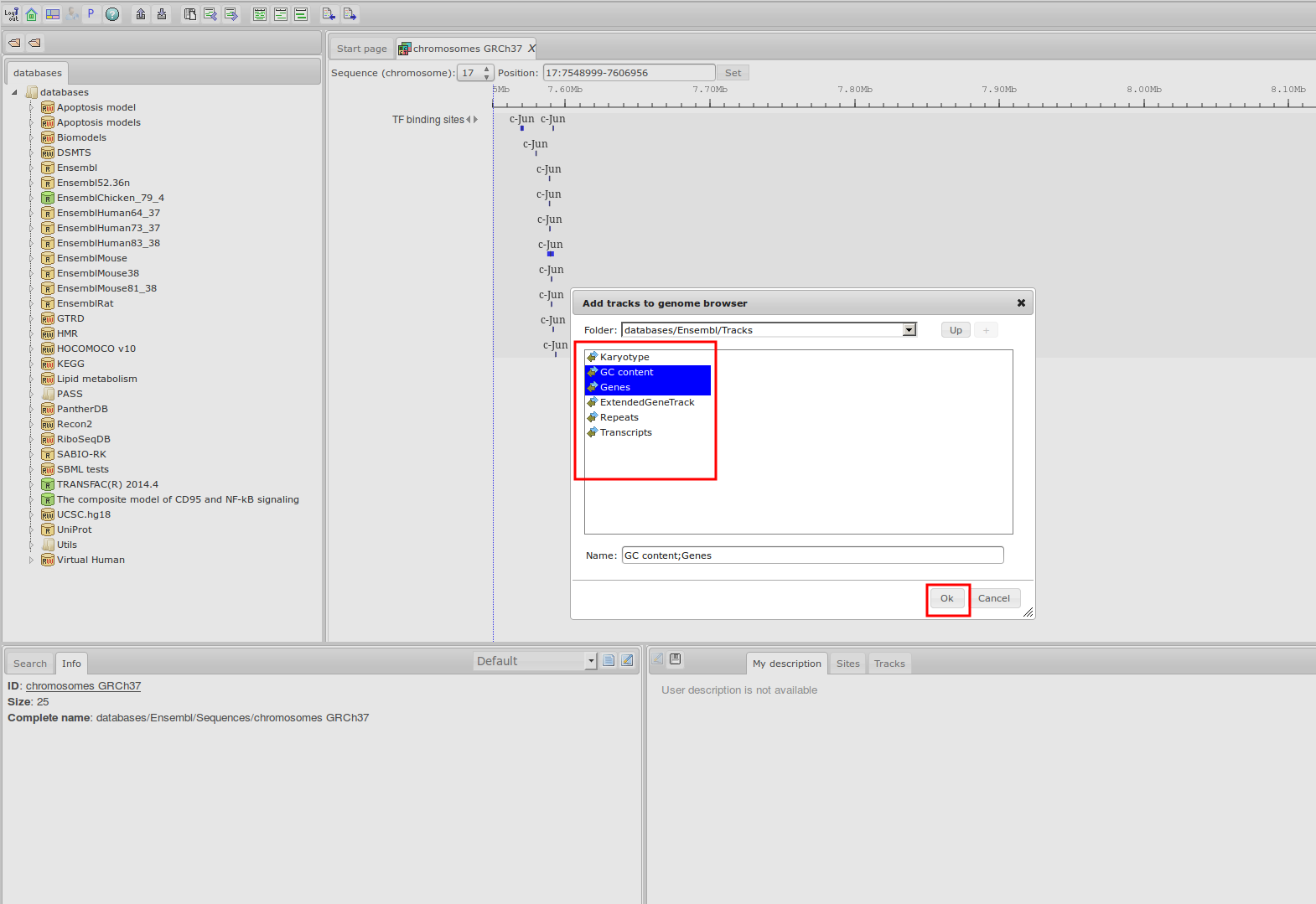
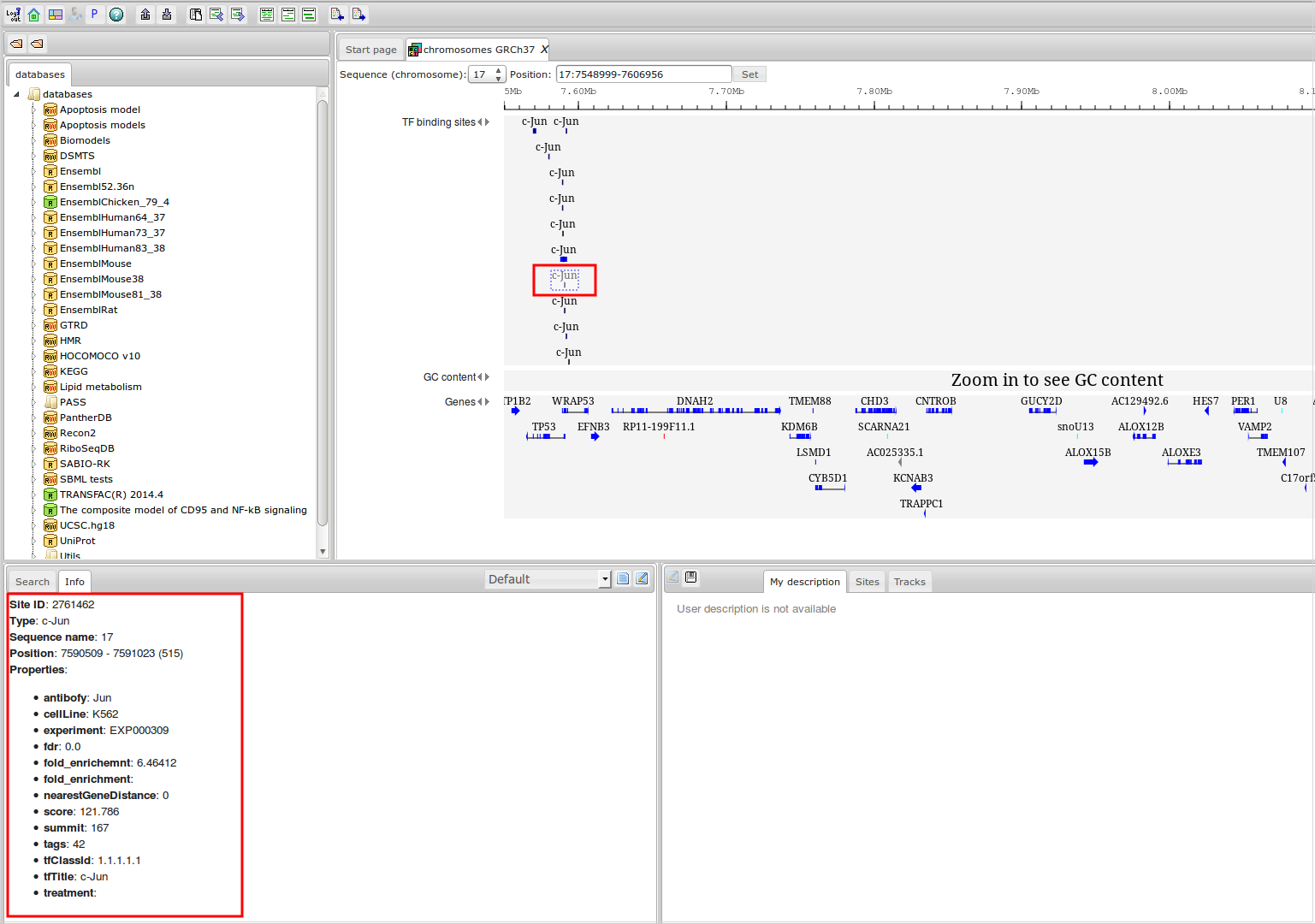
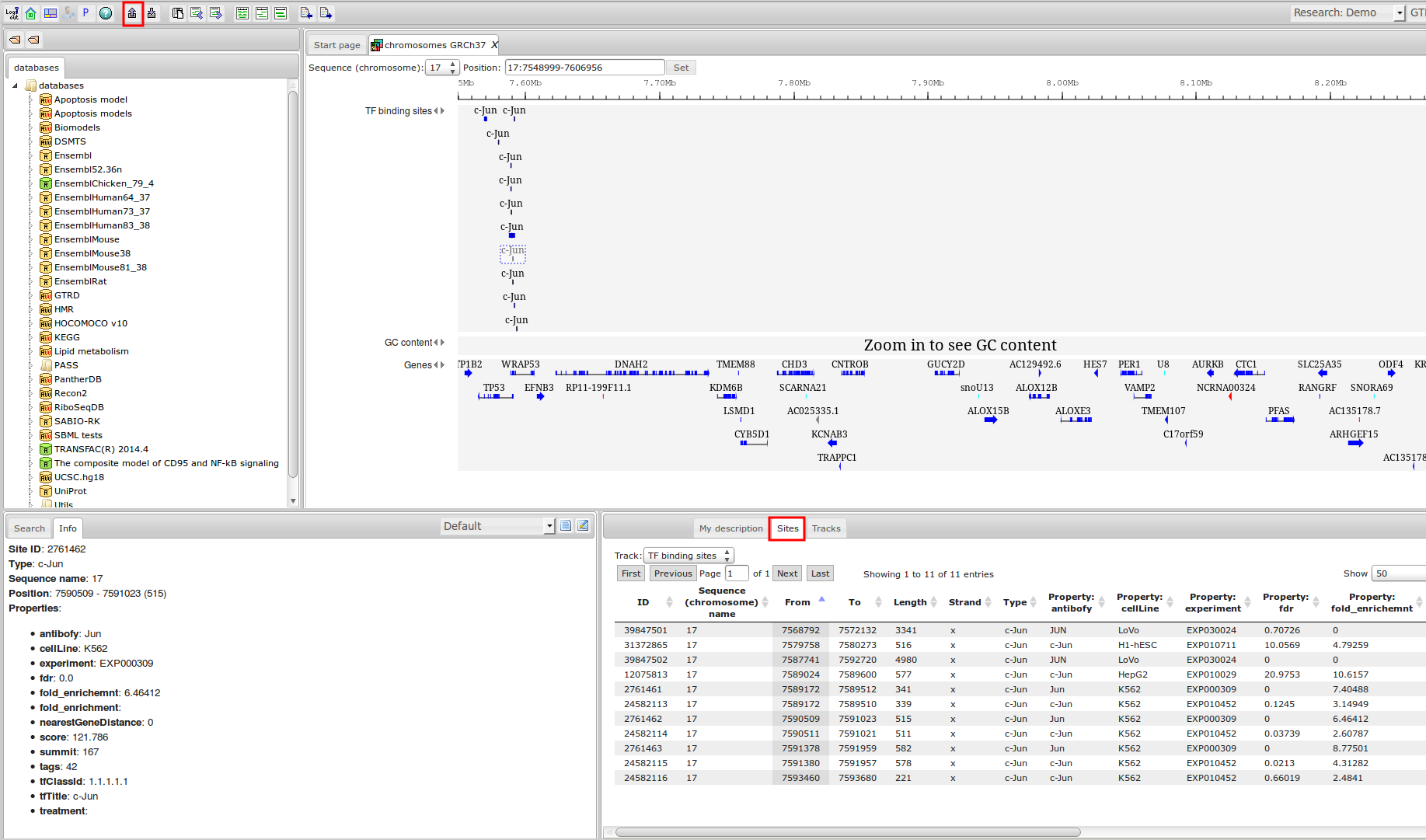
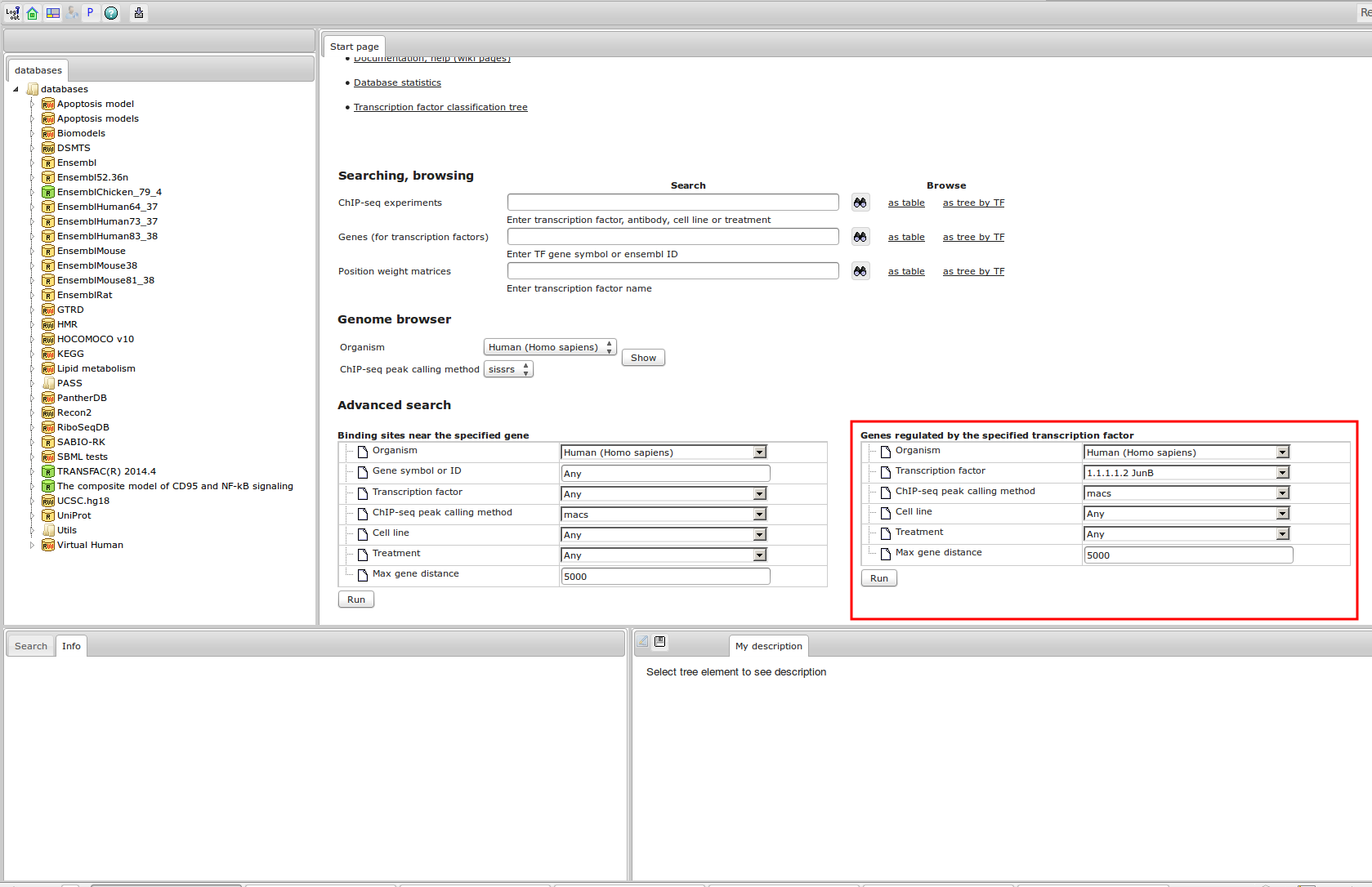
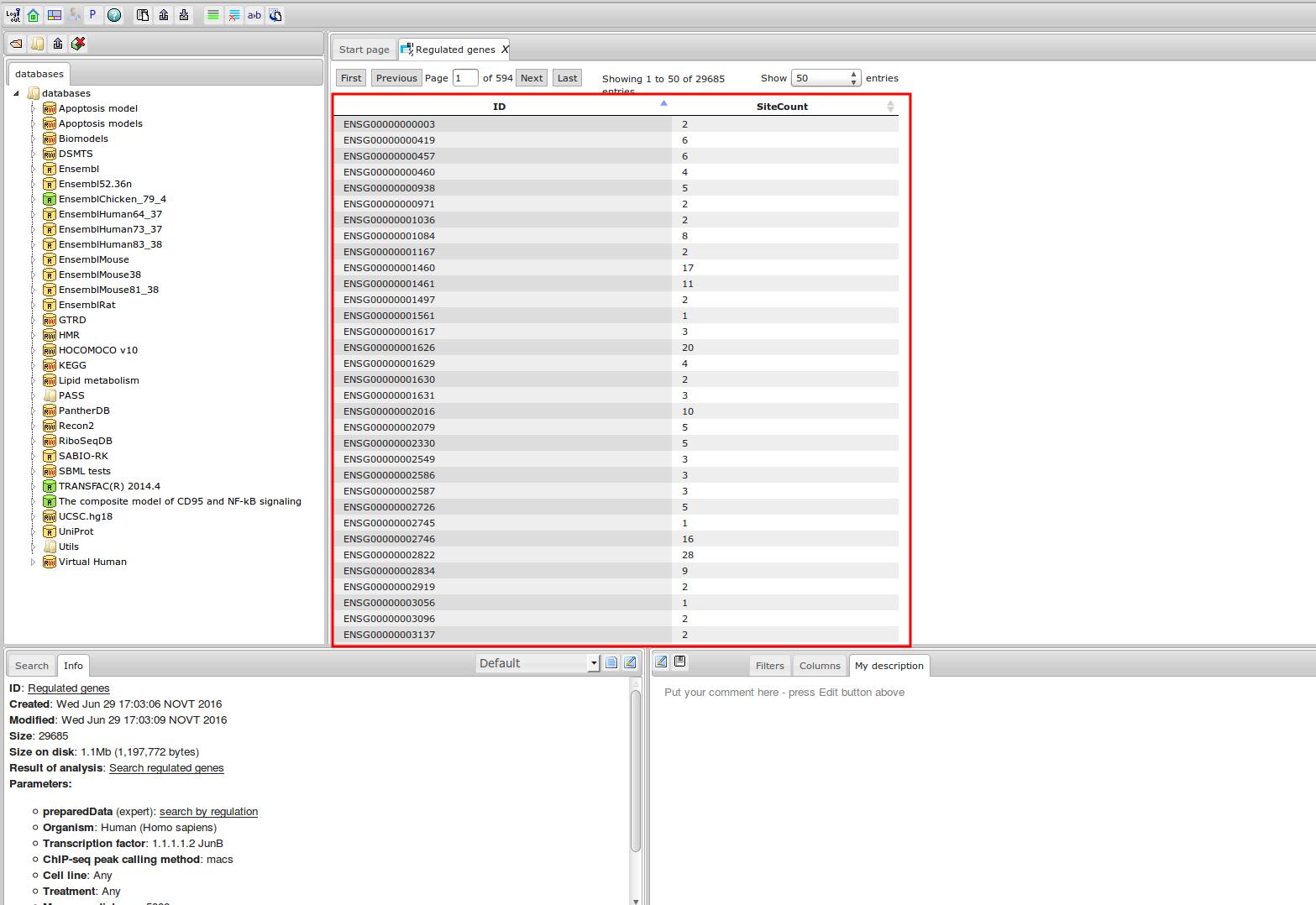
Workflow
How was it constructed?
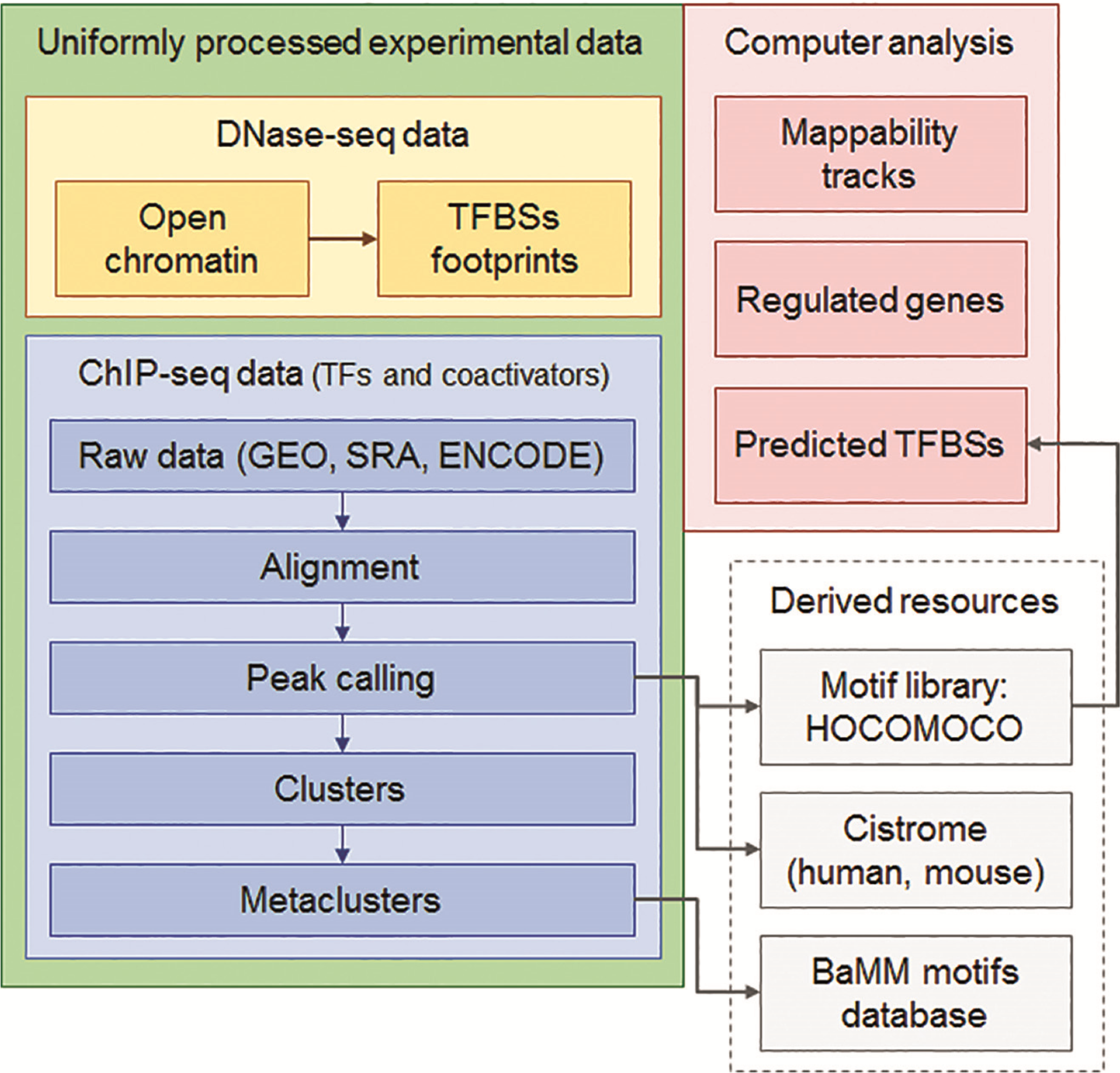
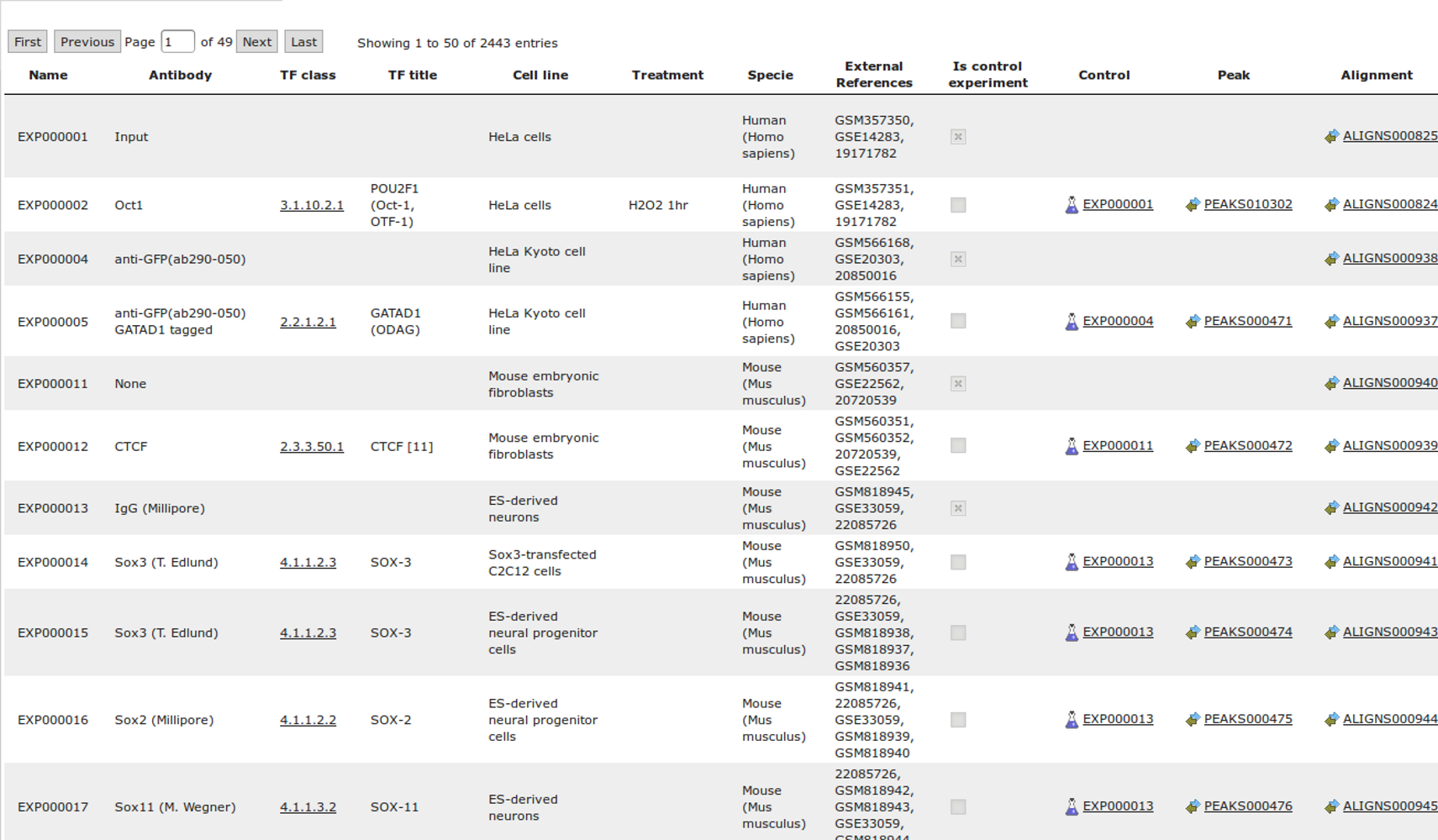
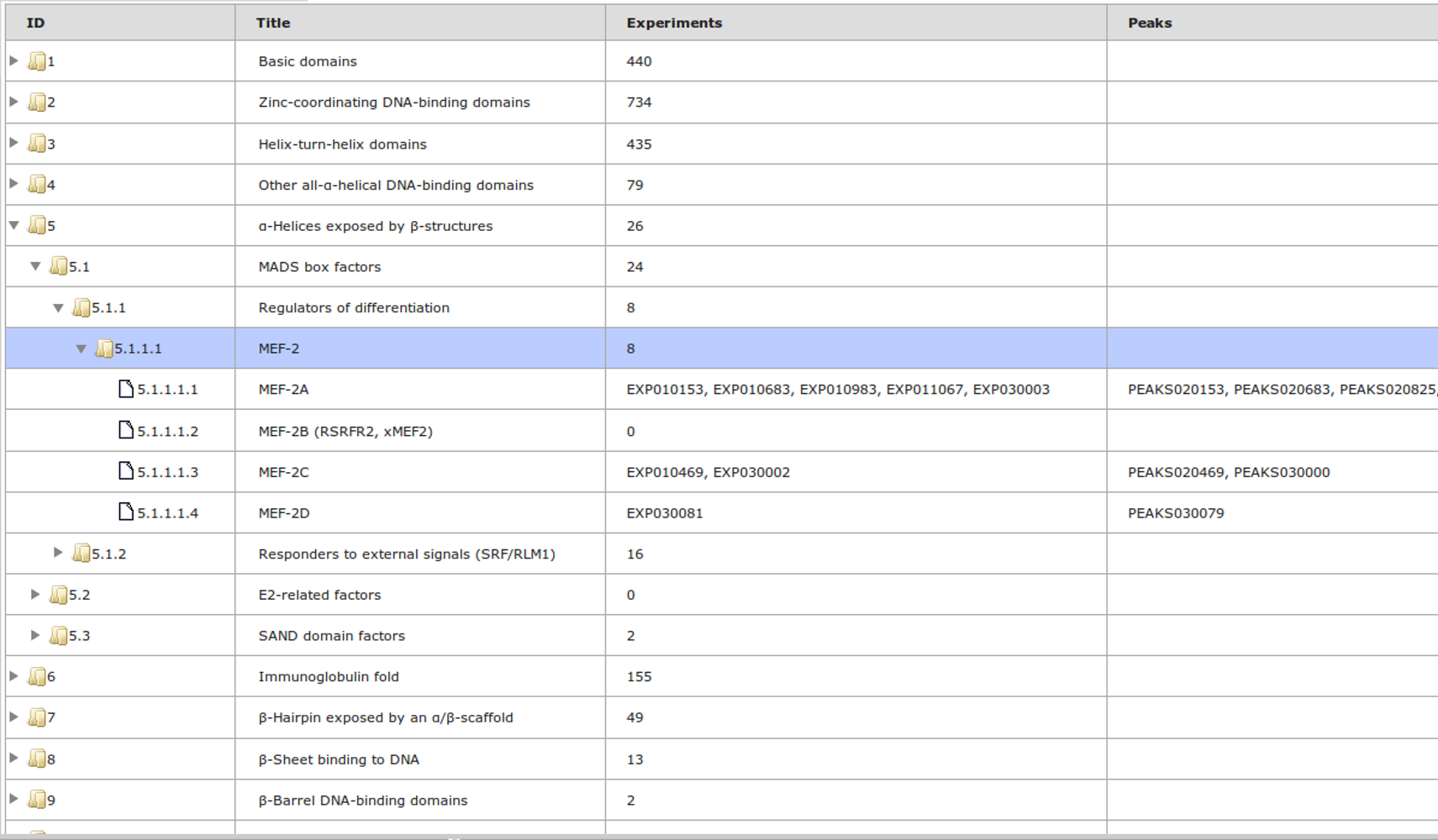
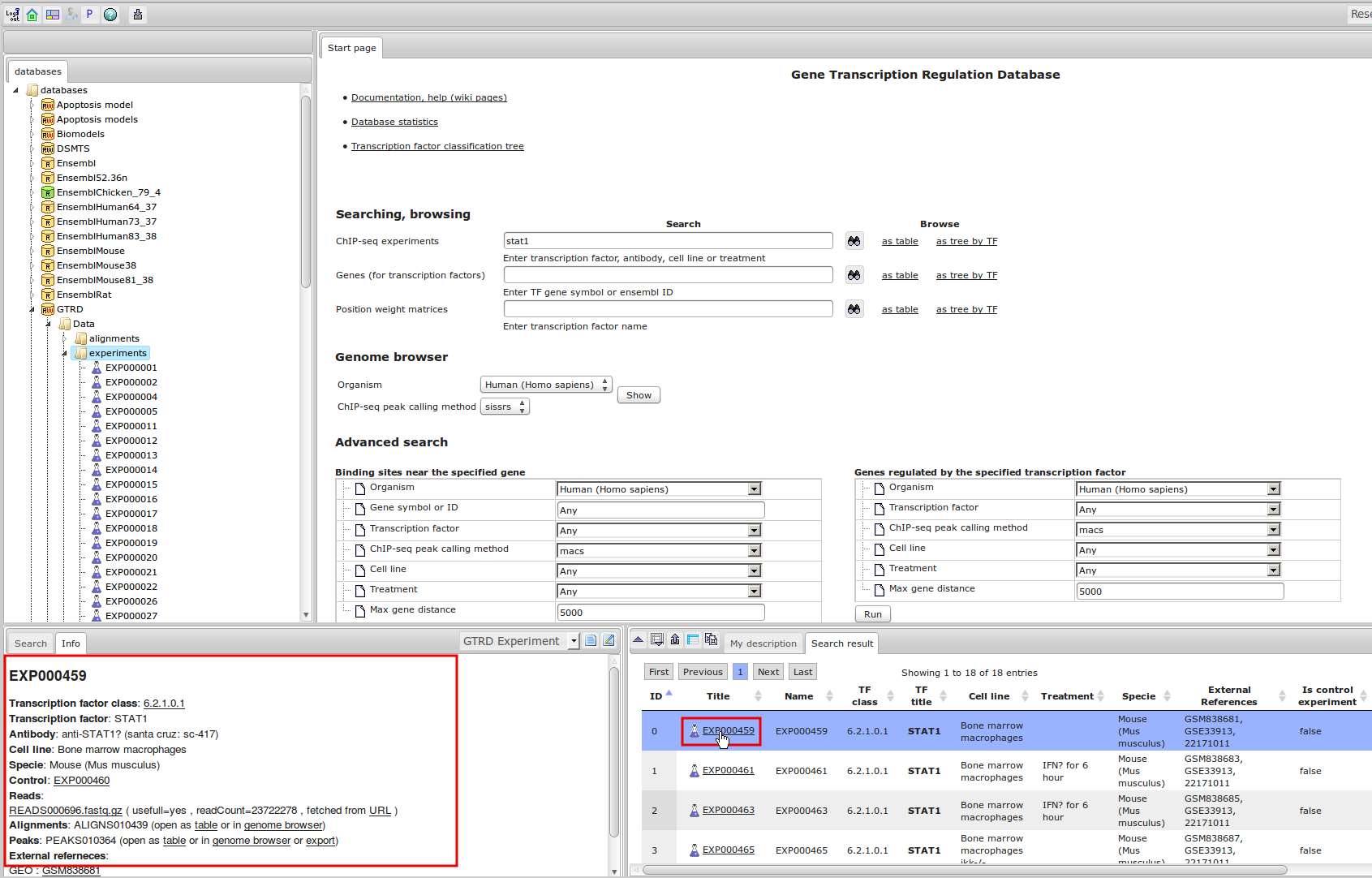
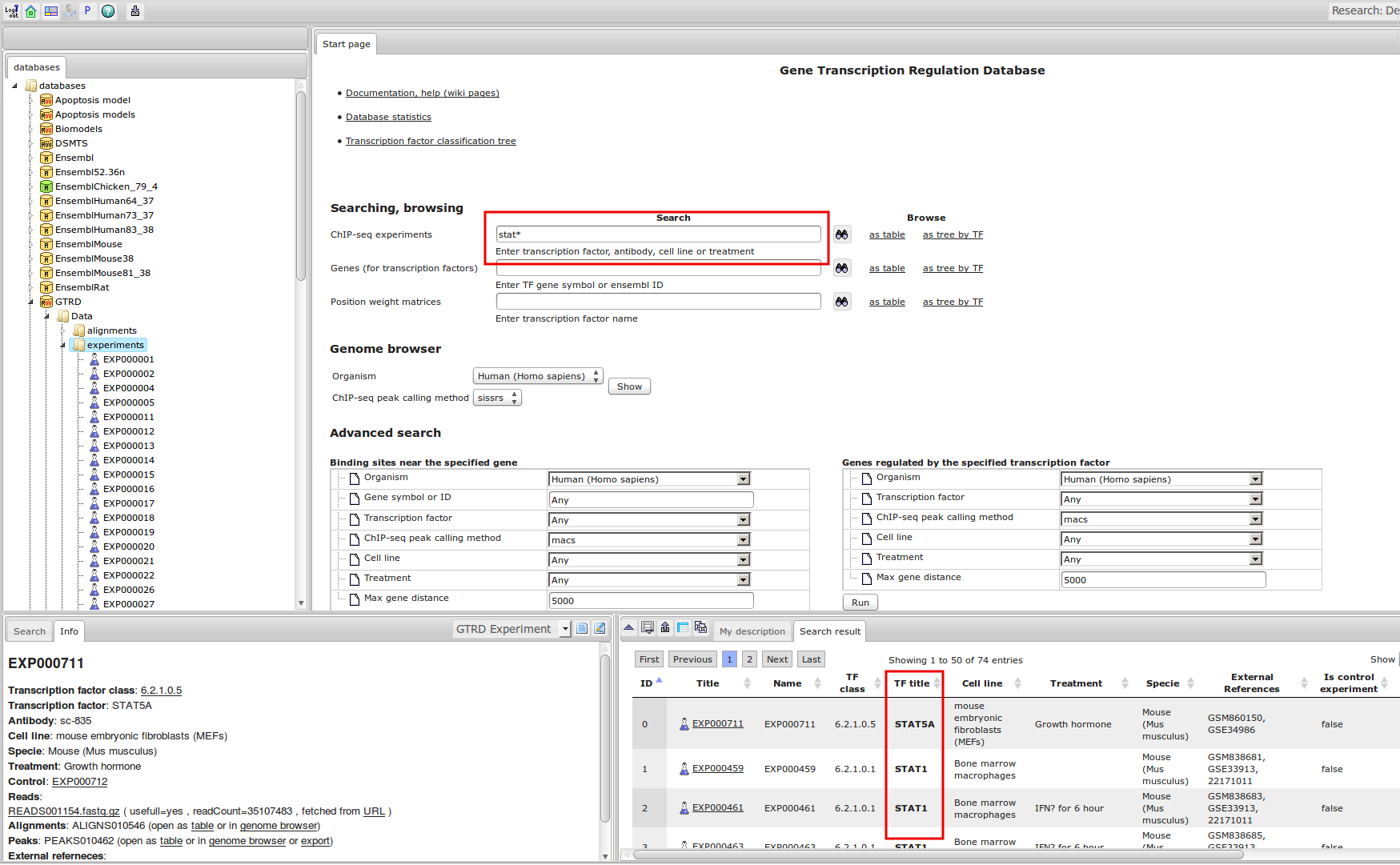
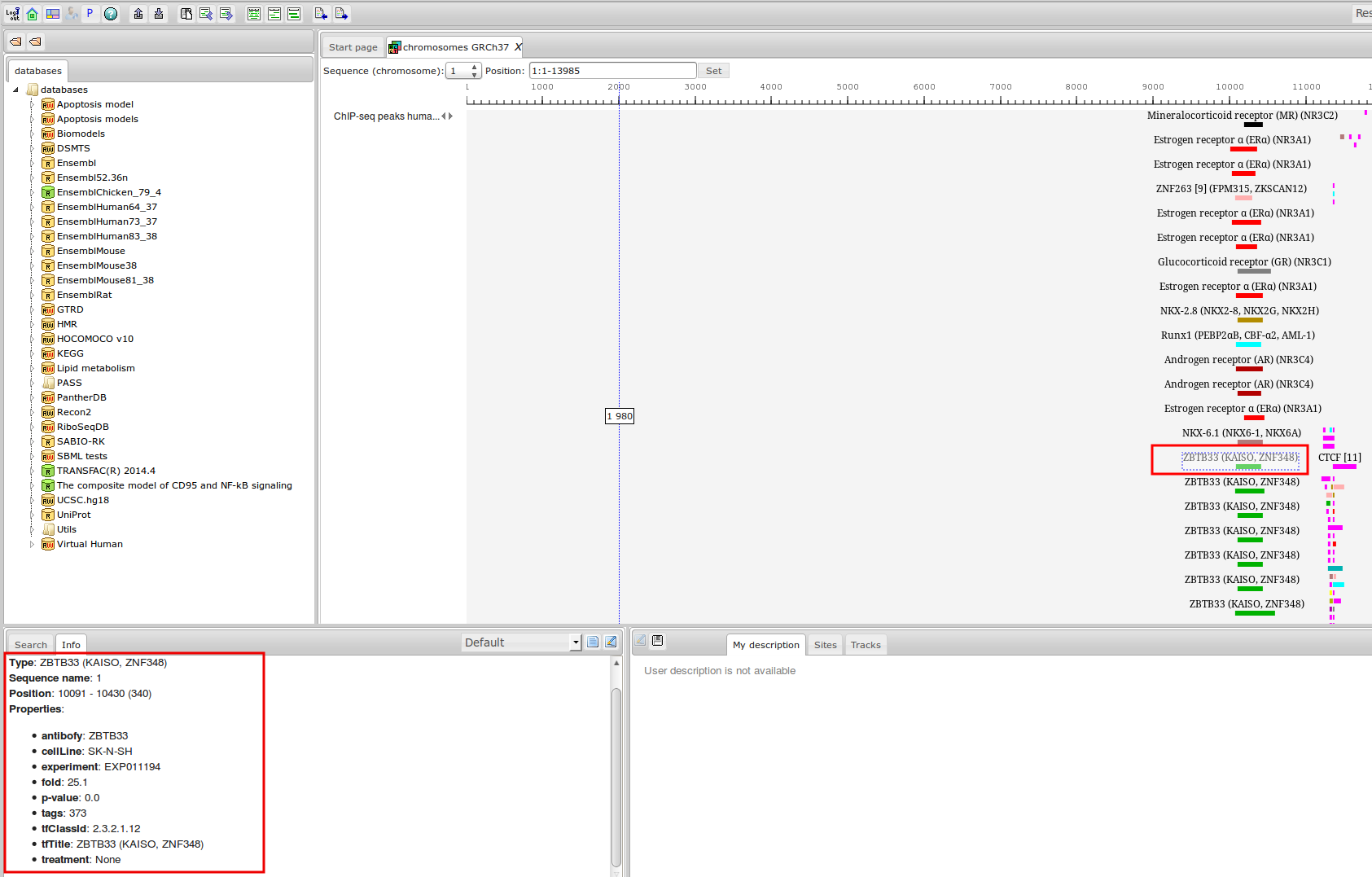
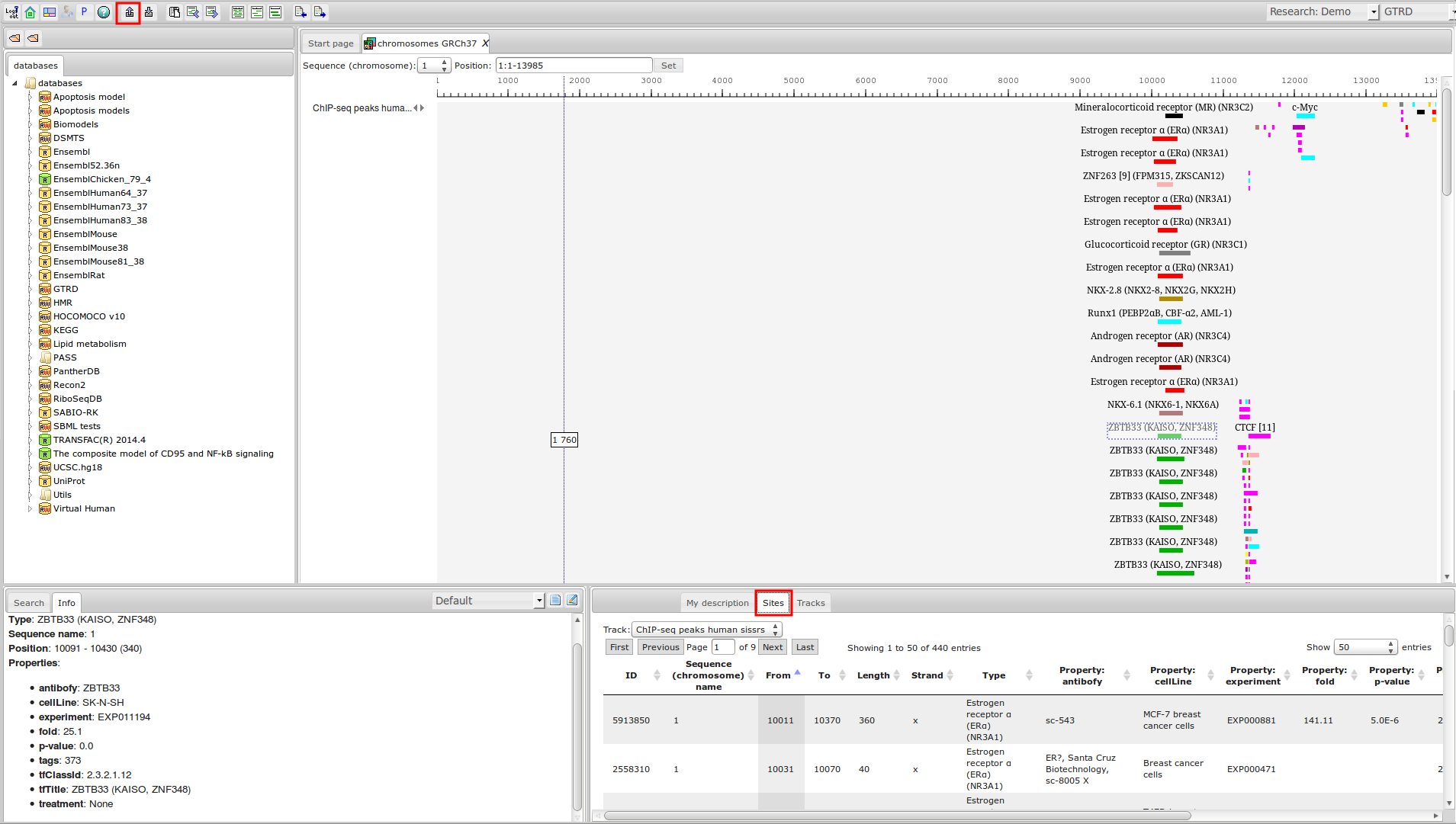
ChIP-seq experiment information and raw data were collected from publically available sources. Sequenced reads were aligned using Bowtie2 and ChIP-seq peaks were called using 4 different methods. Peaks were merged into clusters and metaclusters to produce non-redundant set of transcription factor binding sites.
Learn more »How to cite
License
Users may freely use the GTRD database for non-commercial purposes as long as they properly cite it. If you intend to use GTRD for a commercial purpose, please contact ivan@dote.ru to arrange a license.
Statistics for version 19.04 (version 18.06)
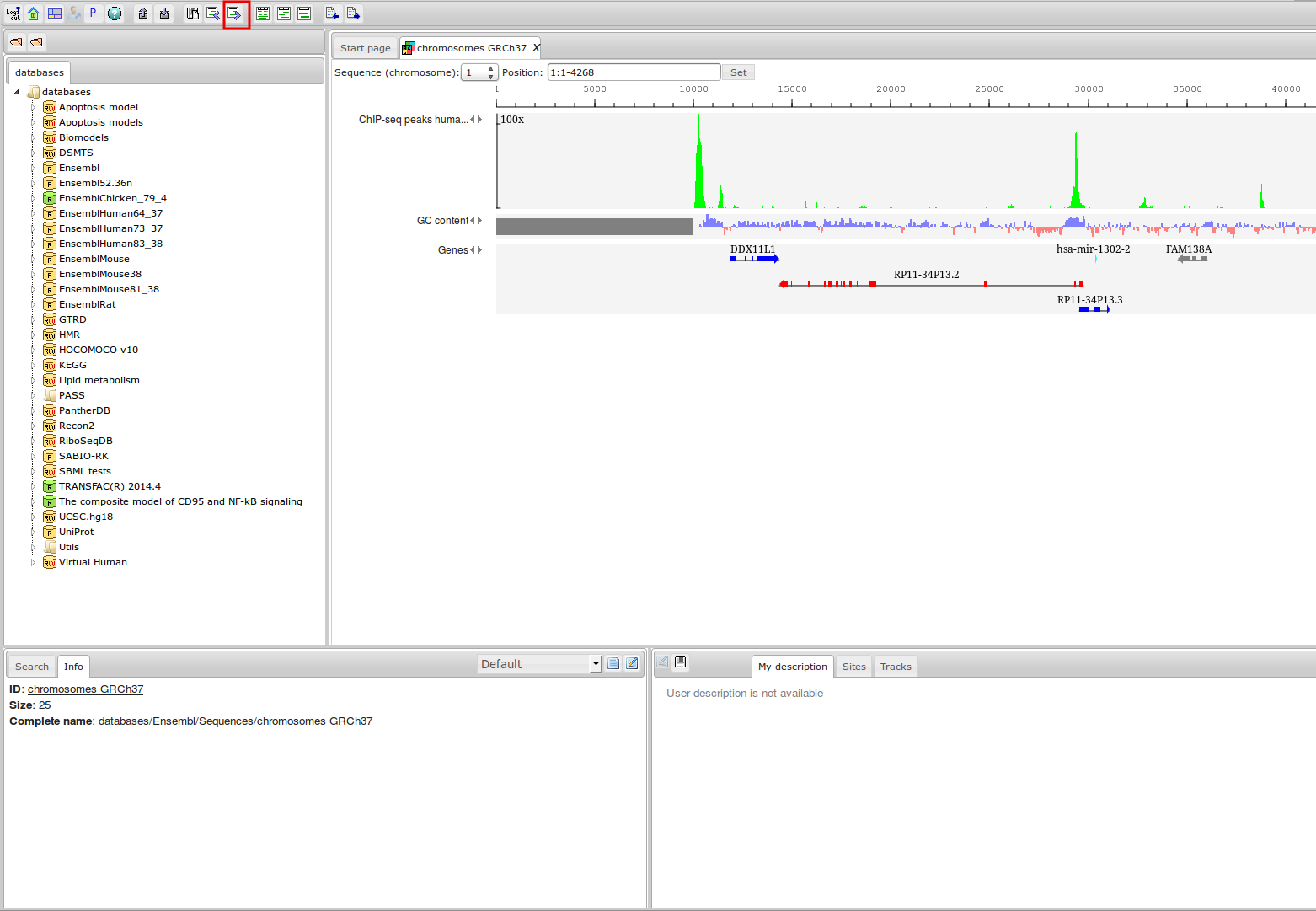
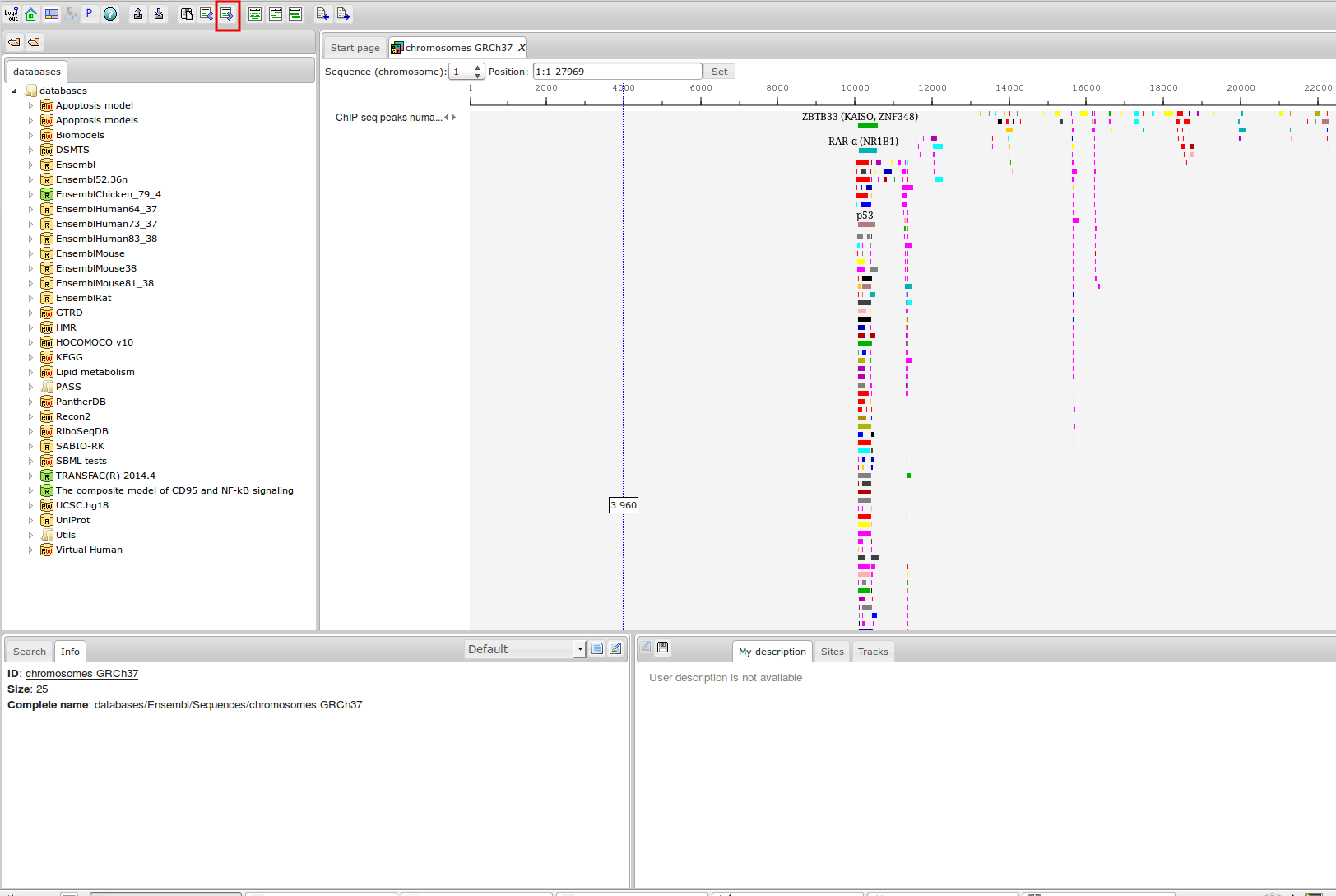
ChIP-seq
| Species | ChIP-seq experiments | Transcription factors | ChIP-seq reads ×109 |
Reads aligned ×109 |
ChIP-seq peaks ×106 |
Clusters ×106 |
Metaclusters ×106 |
|---|---|---|---|---|---|---|---|
| Arabidopsis thaliana | 424 (345) | 120 (105) | 24,2 (19,7) | 14,4 (12,1) | 2,9 (2,8) | 2,2 (2,1) | 0,38 (0,34) |
| Caenorhabditis elegans | 1508 (1264) | 326 (296) | 26,6 (23,7) | 19,1 (16,9) | 3,4 (2,8) | 2,4 (2,0) | 0,4 (0,38) |
| Danio rerio | 51 (51) | 20 (20) | 2,1 (2,1) | 1,1 (1,1) | 2,1 (1,8) | 1,9 (1,7) | 0,26 (0,26) |
| Drosophila melanogaster | 2182 (2016) | 552 (503) | 74,4 (69,9) | 53,1 (49,9) | 10,5 (7,9) | 7,4 (5,6) | 1,4 (1,0) |
| Homo sapiens | 10675 (7239) | 1158 (852) | 616,3 (408,6 ) | 483,3 (321,1) | 818 (551,2) | 505,2 (345,0) | 60,5 (42,7) |
| Mus musculus | 8055 (5521) | 584 (432) | 403,4 (245,7) | 273,1 (173,9) | 699,8 (512,8) | 431,6 (334,1) | 56 (42,9) |
| Rattus norvegicus | 153 (101) | 29 (20) | 7,5 (5,3) | 4,8 (3,2) | 9,3 (6,9) | 7,6 (6,6) | 1,1 (0,86) |
| Saccharomyces cerevisiae | 1364 (777) | 142 (120) | 20,3 (12,3) | 15,5 (10,1) | 0,66 (0,36) | 0,3 (0,22) | 0,038 (0,028) |
| Schizosaccharomyces pombe | 222 (171) | 55 (52) | 3,4 (1,9) | 2,6 (1,4) | 0,11 (0,08) | 0,09 (0,06) | 0,011 (0,009) |
| Total | 24634 (17485) | 2986 (2400) | 1178,3 (789,2) | 867,1 (589,8) | 1546,7 (1086,7) | 958,7 (697,3) | 120,1 (88,6) |
DNase-seq
| Species | DNase-seq experiments | DNase I hypersensitive sites (DHSs) ×106 |
DNase-seq footprints (FDR<0.01) ×106 |
|---|---|---|---|
| Homo sapiens | 1482 (739) |
|
|
| Mus musculus | 103 (103) |
|
|
| Total | 1585 (842) |
|
|
Studies citing GTRD
- ERRα promotes breast cancer cell dissemination to bone by increasing RANK expression in primary breast tumors. Oncogene, 2018 Nov 26
- ChIP-Atlas: a data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep. 2018 Nov 9
- Inference of tumor cell-specific transcription factor binding from cell-free DNA. Preprint. October 2018
- Genome-wide map of human and mouse transcription factor binding sites aggregated from ChIP-Seq data. BMC Res Notes. 2018 Oct 23
- MethMotif: an integrative cell specific database of transcription factor binding motifs coupled with DNA methylation profiles. Nucleic Acids Research. 31 October 2018.
- Characterization of transcriptome transition associates long noncoding RNAs with glioma progression. Molecular Therapy - Nucleic Acids. October 2018.
- An Evolutionarily Conserved Mesodermal Enhancer in Vertebrate Zic3. Scientific Reports. December 2018.
- Introductory Chapter: A Brief Overview of Transcriptional and Post-transcriptional Regulation. In book: Transcriptional and Post-transcriptional Regulation. October 2018.
- Contributions of regulated transcription and mRNA decay to the dynamics of gene expression. Wiley Interdiscip Rev RNA. 2018 Oct.
- A map of direct TF-DNA interactions in the human genome Preprint 2018.
- Disentangling transcription factor binding site complexity. Nucleic Acids Research. August 2018.
- TFmapper : A Tool for Searching Putative Factors Regulating Gene Expression Using ChIP-seq Data. Int J Biol Sci 2018.
- Endolysosomal degradation of Tau and its role in glucocorticoid‐driven hippocampal malfunction. EMBO J. 2018 Oct 15.
- snpEnrichR: analyzing co-localization of SNPs and their proxies in genomic regions. Bioinformatics. 2018 Jun 7.
- Detecting genome-wide directional effects of transcription factor binding on polygenic disease risk. Nature Genetics. September 2018
- Rapid and Efficient FISH using Pre-Labeled Oligomer Probes. Sci Rep. 2018 May 29;8(1):8224.
- Rapamycin Modulates Glucocorticoid Receptor Function, Blocks Atrophogene REDD1, and Protects Skin from Steroid Atrophy. J Invest Dermatol. 2018 Sep;138(9):1935-1944.
- Identifying transcription factor Olig2 genomic binding sites in acutely purified PDGFRα+ cells by low-cell chromatin immunoprecipitation sequencing analysis. J Vis Exp. 2018 Apr 16;(134).
- Functional interplay between c-Myc and Max in B lymphocyte differentiation. EMBO Rep. 2018 Aug 20.
- Genome-Wide Scanning of Gene Expression. Reference Module in Life Sciences. 2018
- Multi-omic Directed Networks Describe Features of Gene Regulation in Aged Brains and Expand the Set of Genes Driving Cognitive Decline Front. Genet., 09 August 2018
- Analysis of diet-induced differential methylation, expression, and interactions of lncRNA and protein-coding genes in mouse liver. Sci Rep. 2018 Aug 1;8(1):11537.
- Systems analysis of dilated cardiomyopathy in the next generation sequencing era. Wiley Interdiscip Rev Syst Biol Med. 2018 Jul
- Hypoxia and serum deprivation induces glycan alterations in triple negative breast cancer cells. Biol Chem. 2018 Jun 27
- Hormone-responsive genes in the SHH and WNT/β-catenin signaling pathways influence urethral closure and phallus growth. Biol Reprod. 2018 May 14.
- The BaMM web server for de-novo motif discovery and regulatory sequence analysis. Nucleic Acids Research, 28 May 2018
- Transcription Factor Databases. Reference Module in Life Sciences. 2018
- Core signaling pathways in ovarian cancer stem cell revealed by integrative analysis of multi-marker genomics data. PLoS One. 2018 May 3.
- Computational master-regulator search reveals mTOR and PI3K pathways responsible for low sensitivity of NCI-H292 and A427 lung cancer cell lines to cytotoxic action of p53 activator Nutlin-3. BMC Medical Genomics, 13 February 2018.
- MnSOD/SOD2 in Cancer: The Story of a Double Agent. Reactive Oxygen Species, Feb 2018.
- The Role of Indoleamine-2,3-Dioxygenase in Cancer Development, Diagnostics, and Therapy. Front. Immunol., 31 January 2018
-
Eukaryotic and prokaryotic promoter databases as valuable tools in exploring the regulation of gene transcription: a comprehensive overview.
Gene. 2017 Nov 2. -
HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis.
Nucleic Acids Res. 2017 Nov 11. - ReMap 2018: an updated atlas of regulatory regions from an integrative analysis of DNA-binding ChIP-seq experiments Nucleic Acids Res. 2017 Nov 8.
- DMS-Seq for In Vivo Genome-wide Mapping of Protein-DNA Interactions and Nucleosome Centers. Cell Reports 2017 Oct 3;21(1):289-300.
- EpiDenovo: a platform for linking regulatory de novo mutations to developmental epigenetics and diseases. Nucleic Acids Research 2017, gkx918
- Genetic variants in ADAMTS13 as well as smoking are major determinants of plasma ADAMTS13 levels. Blood Advances 2017 1:1037-1046;
- Master-regulators driving resistance of non-small cell lung cancer cells to p53 reactivator Nutlin-3. Virtual Biology 2017, 0(4), 1-31.
- Discovering relationships between nuclear receptor signaling pathways, genes, and tissues in Transcriptomine. Sci Signal. 2017 Apr 25;10(476).
- A comprehensive review of web-based non-coding RNA resources for cancer research. Cancer Lett. 2017 Aug 18;407:1-8.
- RUNX1 promote invasiveness in pancreatic ductal adenocarcinoma through regulating miR-93. Oncotarget 2017 Aug 24